Why Choose Biophysics at the University of Maryland?
The Biophysics Graduate Program offers advanced training in theoretical and computational methods in combination with cutting-edge experimental techniques to solve some of the most challenging problems in biology, biomedicine, and bioengineering.

The Biophysics Graduate Program brings together faculty from biochemistry, biology, chemistry, engineering and physics working on questions at the intersection of the physical and life sciences.
The program offers advanced training in theoretical and computational methods in combination with cutting-edge experimental techniques to solve some of the most challenging problems in biology, biomedicine, and bioengineering.

Program participants have full access to on-campus NMR, SAXS, imaging and microfabrication facilities as well as several high-performance computing clusters.
The proximity to NIH and NIST provides unlimited possibilities for collaborations and use of unique equipment.

Many of our students are awarded fellowships through the NCI-UMD Partnership and participate in the NSF-funded Computation and Mathematics for Biological Networks (COMBINE) and the Physics of Living Systems Network (PoLS) programs.
We provide financial aid via Teaching Assistantships and Dean's Fellowships to all entering graduate students. Funding includes tuition remission and health insurance benefits. Outstanding students are automatically considered for University-wide Fellowships and may be offered Research Assistantships.

Our graduates go on to successful careers in academia, government, and industry.
Meet our current graduate students and check out what they're up to.
Recent Student Publications
Riya Samanta
First-author paper in Biophysical Journal
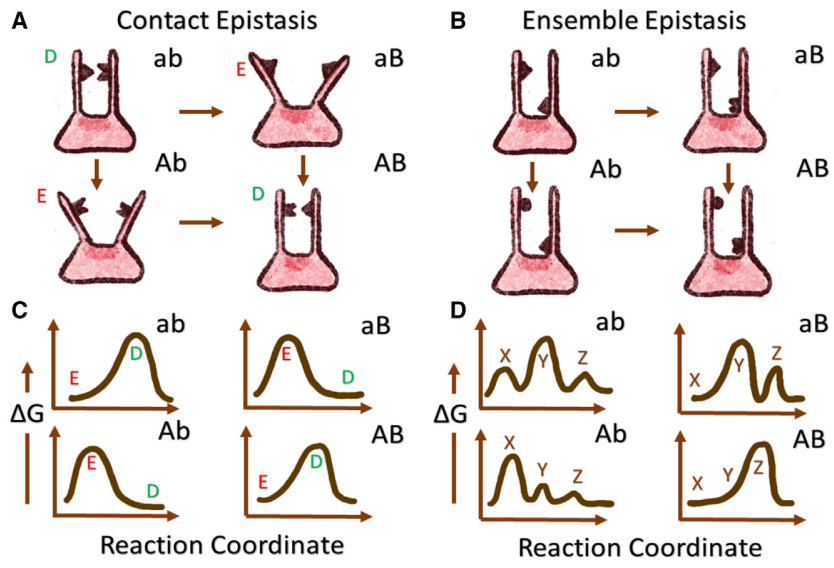
"Decoupling epistasis mechanisms in biomacromolecules"
Zachary Smith
Second-author paper in Angewandte Chemie
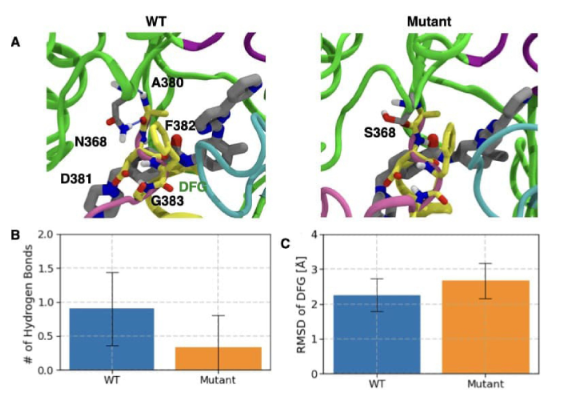
"Protein Flexibility and Dissociation Pathway Differentiation Can Explain Onset of Resistance Mutations in Kinases"
Chenghang Zhang
First-author paper in Cell Reports
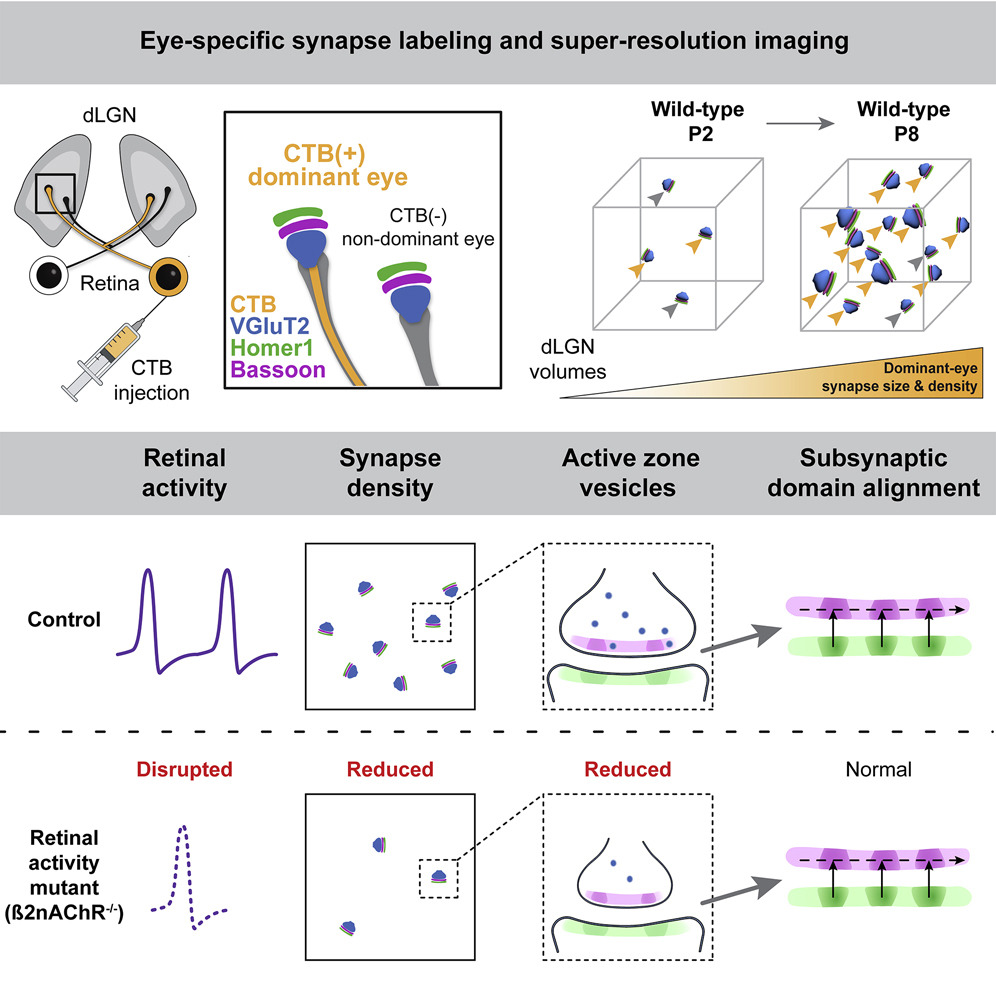
"The synaptic basis of activity-dependent eye-specific competition"
Akashnathan Aranganathan
Second-author paper in the Journal of Chemical Theory and Computation
"AlphaFold2-RAVE: From Sequence to Boltzmann Ranking"